1. Introduction
Plant hormones play an important role as signaling molecules in the regulation of growth and development by controlling the expression of downstream target genes. Furthermore, hormone signaling is a complex network involving interaction of and regulation by other phytohormones. Thus, a comprehensive and integrative analysis of hormone and gene expression will contribute to the understanding of the overall mechanism of action of plant hormones. This database provides an integrated data set of hormonome and transcriptome analyses in 14 organs of the rice plant at the reproductive stage and in gibberellin-related mutants. Users can easily search objective data by using plant hormone names, probe IDs, locus IDs, and gene descriptions as queries. The data of transcripts and hormones are easily visualized as heat maps.
2. Materials and Methods
2-1. Rice plant organs
Oryza sativa L. cv. Nipponbare was grown in soil in a greenhouse with irrigation and supplemental artificial light. At the heading stage, the plant organs shown in Table 1 were harvested and immediately frozen in liquid nitrogen after measurement of fresh weight. The harvested tissues were stored at -80°C until extraction of phytohormones or total RNA.
2-2. GA mutants
Growth and harvest conditions of GA mutants (Table 1) were described in a previous paper (Kojima et al. 2009).
| Nos. | Plant organs | Abbreviations | Biological replicate | |
| GeneChip | Hormone | |||
| Organs at the heading stage | ||||
| 1 | Flowers before anthesis | Flw | 3 | 3 |
| 2 | Panicle branches | PBr | 3 | 3 |
| 3 | Top part of internode I | InN I-top | 3 | 3 |
| 4 | Basal part of internode I | InN I-bsl | 3 | 3 |
| 5 | Node I | Nod I | 3 | 3 |
| 6 | Node II | Nod II | 3 | 3 |
| 7 | Tip of the blade of the flag leaf | FLB-tip | 3 | 3 |
| 8 | Middle part of the blade of the flag leaf | FLB-mid | 3 | 3 |
| 9 | Basal part of the blade of the flag leaf | FLB-bsl | 3 | 3 |
| 10 | Top part of the sheath of the flag leaf | FLS-top | 3 | 3 |
| 11 | Basal part of the sheath of the flag leaf | FLS-bsl | 2 | 3 |
| 12 | Whole blade of the flag leaf | FLB | 1 | 3 |
| 13 | Whole blade of leaf 2 counted down from the flag leaf | LB-2 | 1 | 3 |
| 14 | Whole blade of leaf 4 counted down from the flag leaf | LB-4 | 1 | 3 |
| Gibberellin-related mutants | ||||
| Shoot of Taichung 65 (control) | T65 | 3 | 3 | |
| Shoot of gid1-3 mutant | gid1 | 3 | 3 | |
| Shoot of gid2-1 mutant | gid2 | 3 | 3 | |
| Shoot of slr1 mutant | sir1 | 3 | 3 | |
2-3. Phytohormone analysis
Phytohormone extraction, purification, and quantification were carried out as described previously (Kojima et al. 2009).
| Hormone | Class | Abbreviation | Physiological property |
| Abscisic acid | Abscisic acid | ABA | Active compound |
| Indole-3-acetic acid | Auxin | IAA | Active compound |
| Indole-3-acetyl-L-alanine | Auxin | IAAla | Deactivated compound |
| Indole-3-acetyl-L-isoleucine | Auxin | IAIle | Deactivated compound |
| Indole-3-acetyl-L-leucine | Auxin | IALeu | Deactivated compound |
| Indole-3-acetyl-L-aspartic acid | Auxin | IAAsp | Deactivated compound |
| Indole-3-acetyl-L-tryptophan | Auxin | IATrp | Deactivated compound |
| Indole-3-acetyl-L-phenylalanine | Auxin | IAPhe | Deactivated compound |
| N6-(ÃÂ2-Isopentenyl)adenine | Cytokinin | iP | Active compound |
| N6-(ÃÂ2-Isopentenyl)adenine riboside | Cytokinin | iPR | Intermediate |
| N6-(ÃÂ2-Isopentenyl)adenine ribotides | Cytokinin | iPRPs | Intermediate |
| N6-(ÃÂ2-Isopentenyl)adenine-N7-glucoside | Cytokinin | iP7G | Deactivated compound |
| N6-(ÃÂ2-Isopentenyl)adenine-N9-glucoside | Cytokinin | iP9G | Deactivated compound |
| trans-Zeatin | Cytokinin | tZ | Active compound |
| trans-Zeatin riboside | Cytokinin | tZR | Intermediate |
| trans-Zeatin ribotides | Cytokinin | tZRPs | Intermediate |
| trans-Zeatin-N7-glucoside | Cytokinin | tZ7G | Deactivated compound |
| trans-Zeatin-N9-glucoside | Cytokinin | tZ9G | Deactivated compound |
| trans-Zeatin-O-glucoside | Cytokinin | tZOG | Deactivated compound |
| trans-Zeatin riboside-O-glucoside | Cytokinin | tZROG | Deactivated compound |
| trans-Zeatin ribotide-O-glucosides | Cytokinin | tZRPsOG | Deactivated compound |
| cis-Zeatin | Cytokinin | cZ | Active compound |
| cis-Zeatin riboside | Cytokinin | cZR | Intermediate |
| cis-Zeatin ribotides | Cytokinin | cZRPs | Intermediate |
| cis-Zeatin-O-glucoside | Cytokinin | cZOG | Deactivated compound |
| cis-Zeatin riboside-O-glucoside | Cytokinin | cZROG | Deactivated compound |
| cis-Zeatin ribotide-O-glucosides | Cytokinin | cZRPsOG | Deactivated compound |
| Dihydro-zeatin | Cytokinin | DZ | Active compound |
| Dihydro-zeatin riboside | Cytokinin | DZR | Intermediate |
| Dihydro-zeatin ribotides | Cytokinin | DZRPs | Intermediate |
| Dihydro-zeatin-N9-glucoside | Cytokinin | DZ9G | Deactivated compound |
| Gibberellin A1 | Gibberellin | GA1 | Active compound |
| Gibberellin A3 | Gibberellin | GA3 | Active compound |
| Gibberellin A4 | Gibberellin | GA4 | Active compound |
| Gibberellin A7 | Gibberellin | GA7 | Intermediate |
| Gibberellin A8 | Gibberellin | GA8 | Deactivated compound |
| Gibberellin A9 | Gibberellin | GA9 | Intermediate |
| Gibberellin A12 | Gibberellin | GA12 | Intermediate |
| Gibberellin A19 | Gibberellin | GA19 | Intermediate |
| Gibberellin A20 | Gibberellin | GA20 | Intermediate |
| Gibberellin A24 | Gibberellin | GA24 | Intermediate |
| Gibberellin A44 | Gibberellin | GA44 | Intermediate |
| Gibberellin A53 | Gibberellin | GA53 | Intermediate |
2-4. Transcriptomic analysis
Microarray analysis was performed using a GeneChip® Rice Genome Array (Affymetrix). Extraction of total RNA was carried out by using RNeasy® Mini Kit (QIAGEN). Preparation of labeled target cRNA and the subsequent purification and fragmentation of cRNA were carried out using Genechip® One-Cycle Target Labeling and Control Reagents (Affymetrix). Double-stranded cDNA was prepared from 5 µg of total RNA. Hybridization, washing, staining, and scanning were performed as described in the supplierâÂÂs protocol. A 5-µg aliquot of fragmented cRNA was used for hybridization to microarrays. These experiments were conducted according to the manufacturer's guidelines.
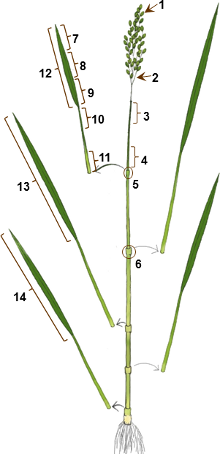
Figure 1. Illustration of rice plant organs used for hormone and transcriptome analyses. 1, Flowers before anthesis; 2, Panicle branches; 3, Top part of internode I; 4, Basal part of internode I; 5, Node I; 6, Node II; 7, Tip of the blade of the flag leaf; 8, Middle part of the blade of the flag leaf; 9, Basal part of the blade of the flag leaf; 10, Top part of the sheath of the flag leaf; 11, Basal part of the sheath of the flag leaf; 12, Whole blade of the flag leaf; 13, Whole blade of leaf 2 counted down from the flag leaf; 14, Whole blade of leaf 4 counted down from the flag leaf. The numbers correspond to the numbers shown in Table 1.
3. References
- Kojima, M., Kamada-Nobusada, T., Komatsu, H., Takei, K., Kuroha, T., Mizutani, M., Ashikari, M., Ueguchi-Tanaka, M., Matsuoka, M., Suzuki, K., and Sakakibara, H. (2009) Highly-sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol. 50: 1201-1214. [PubMed]
4. Raw Data
4-1. Phytohormone analysis
- Rice-hormone organs reproductive [46 KB Microsoft Excel 2007 format (zip compressed)]
4-2. Transcriptomic analysis
- Expression data from rice organs at the reproductive stage [NCBI Gene Expression Omnibus]
- Transcriptome analysis of gibberellin-signaling mutants in rice [Oryza sativa] [NCBI Gene Expression Omnibus]
5. Contact Person
- Hitoshi SAKAKIBARA
- E-mail: sakaki at riken jp
- Plant Productivity Systems Research Group,
Plant Science Center, RIKEN